RNA structure prediction
sampling & clustering
A RiboNucleic Acid (RNA) can form complex structure through intra-molecular base-pairing. Some classes of RNAs can regulate biological functions by changing its conformations. An example is illustrated below.
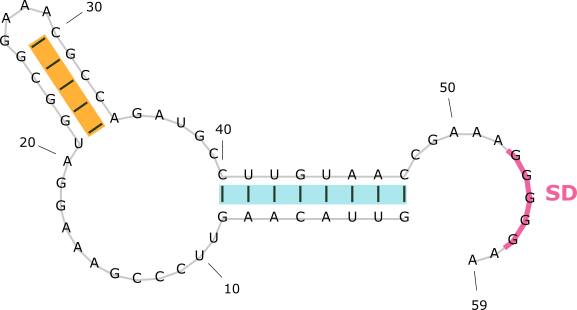
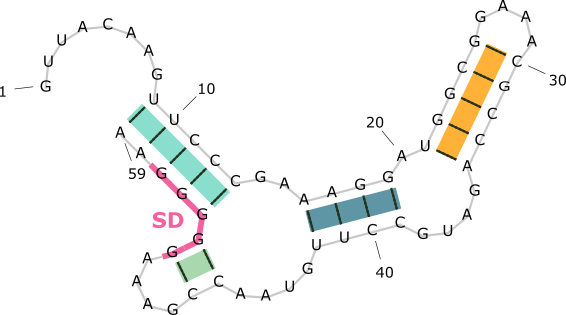
The SAM-III riboswitch folds into two distinct secondary structures. It regulates gene expression by exposing or sequestering the SD sequence, which controls translation.
Identifying multiple structures of a RNA can bring therapeutic advancements for RNA viruses. A popular approach is to sample low-energy structures from the nearest neighbor thermodyanmic model. Most algorithms follow the general flow of sampling, clustering, and reporting cluster representatives.
I worked on improving the clustering aspect of an RNA structure prediction algorithm called profiling. The current method resulted in too many clusters with negligible biological difference. I proposed algorithmic ways to identify clusters that should be merged based on structural similarity. The enhanced version of profiling is under development by Georgia Tech Discrete Mathematics and Molecular Biology group.
I also examined the prospect of using current methods to identify new multimodal RNAs. I found that there is a class of RNAs (kinetic riboswitches) that is difficult to detect from current sampling methods. I proposed a simple co-transcription simulation method to identify multimodality of such RNAs. The results have been published in this paper.
Georgia Tech (2018-2019), joint work with Christine Heitsch (Georgia Tech) and Alain Laederach (UNC).